Immersive Visualization / IQ-Station Wiki
This site hosts information on virtual reality systems that are geared toward scientific visualization, and as such often toward VR on Linux-based systems. Thus, pages here cover various software (and sometimes hardware) technologies that enable virtual reality operation on Linux.
The original IQ-station effort was to create low-cost (for the time) VR systems making use of 3DTV displays to produce CAVE/Fishtank-style VR displays. That effort pre-dated the rise of the consumer HMD VR systems, however, the realm of midrange-cost large-fishtank systems is still important, and has transitioned from 3DTV-based systems to short-throw projectors.
Difference between revisions of "VRVolVis"
m (Added PBRT references) |
(Update of recent accomplishments with ANARI and ANARI backends) |
||
| Line 6: | Line 6: | ||
* R,G&B volumetric (vector) data, | * R,G&B volumetric (vector) data, | ||
* rendering in real time for virtual reality (VR). | * rendering in real time for virtual reality (VR). | ||
==Recent Results== | |||
(Placed at the top for quick review) | |||
===Some recent development highlights:=== | |||
* At least one (perhaps two) ANARI backends now properly handle multiple overlapping volume rendering | |||
** The [[Visionaray]] (<TT>visionaray_cuda</TT>) ANARI backend has been tested — images below | |||
** The [[Barney]] ANARI backend has reportedly been fixed, but has not (yet) been confirmed — will do so by July 12 | |||
* The red,green,blue test volumes have been replaced by actual 3D microscope data (skin) provided by the Mayo Clinic. This is a new dataset, but generally similar to the original (a bit larger). | |||
* ANARI's "remote" rendering backend feature has been tested, and presently works in some circumstances, but not yet for the volume rendering. | |||
** The author of the "remote" backend is currently refactoring the code, and my test application is being used as the Guinea Pig. Rapid progress is being made, so we hope to have this working by July 17. | |||
===Next on the list=== | |||
* Test the [[Barney]] ANARI backend for overlapping volume rendering | |||
* Continue working on testing/updating the "remote" rendering ANARI backend | |||
** NOTE: one concern is how to access data local to the remote machine for the volumes — otherwise the data would have to be sent through the socket connection which would be problematic for large datasets | |||
* Update the ANARI interface to ILLIXR, and add the Mayo Clinic volume rendering as an example application | |||
* Work on new ANARI camera that can render a 360 image over the "remote" interface | |||
* Begin developing a user interface that allows the user to select "blobs" as nuclei, etc. | |||
* Work on an OpenXR interface | |||
* Explore ANARI rendering via a WebGL interface | |||
===Some pictures=== | |||
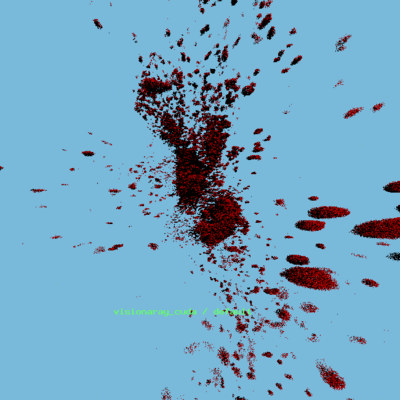
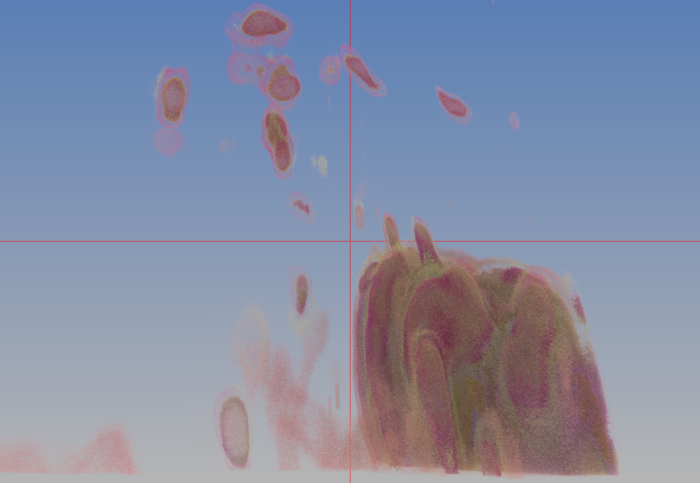
* Toirt Samhliagh rendering of new data — red-channel only | |||
[[File:Toirt_1.png|400px]] | |||
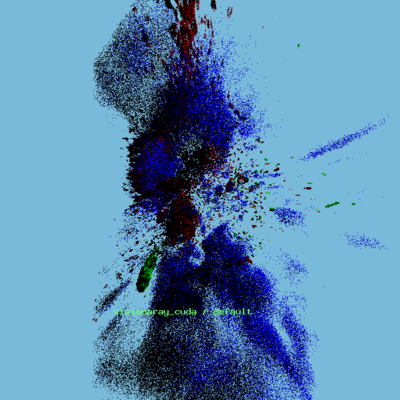
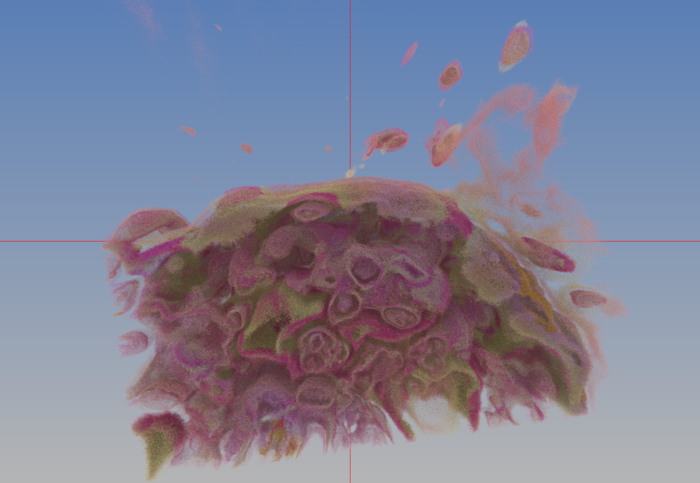
* ANARI-VR rendering of new data — red-channel only | |||
[[File:Ex9c_9.png|400px]] | |||
* ANARI-VR rendering of new data — three-channel | |||
[[File:Ex9c_10.png|400px]] | |||
==Data== | ==Data== | ||
Revision as of 13:28, 11 July 2024
VR VolViz
This page describes various file formats, file conversion techniques and software that can be used to manipulate and render 3D volumes of data using volume-rendering techniques. Much of what is described here will easily work with single-scalar volumetric data, but challenges arise when there is a need for
- near-terabyte sized data,
- R,G&B volumetric (vector) data,
- rendering in real time for virtual reality (VR).
Recent Results
(Placed at the top for quick review)
Some recent development highlights:
- At least one (perhaps two) ANARI backends now properly handle multiple overlapping volume rendering
- The Visionaray (visionaray_cuda) ANARI backend has been tested — images below
- The Barney ANARI backend has reportedly been fixed, but has not (yet) been confirmed — will do so by July 12
- The red,green,blue test volumes have been replaced by actual 3D microscope data (skin) provided by the Mayo Clinic. This is a new dataset, but generally similar to the original (a bit larger).
- ANARI's "remote" rendering backend feature has been tested, and presently works in some circumstances, but not yet for the volume rendering.
- The author of the "remote" backend is currently refactoring the code, and my test application is being used as the Guinea Pig. Rapid progress is being made, so we hope to have this working by July 17.
Next on the list
- Test the Barney ANARI backend for overlapping volume rendering
- Continue working on testing/updating the "remote" rendering ANARI backend
- NOTE: one concern is how to access data local to the remote machine for the volumes — otherwise the data would have to be sent through the socket connection which would be problematic for large datasets
- Update the ANARI interface to ILLIXR, and add the Mayo Clinic volume rendering as an example application
- Work on new ANARI camera that can render a 360 image over the "remote" interface
- Begin developing a user interface that allows the user to select "blobs" as nuclei, etc.
- Work on an OpenXR interface
- Explore ANARI rendering via a WebGL interface
Some pictures
- Toirt Samhliagh rendering of new data — red-channel only
- ANARI-VR rendering of new data — red-channel only
- ANARI-VR rendering of new data — three-channel
Data
Much of the experimental work described here is based on a volumetric dataset created by a 3D microscope, which produces real-color images stacked into the volume. That dataset is too large to provide for quick downloads, so an alternative source for example datasets is provided here (though most are the standard single-scalar type).
Software
Software that has been tested with this dataset (though often with some method of size reduction employed) include:
- ParaView
- ANARI
- VisRTX ANARI backend
- Visionaray ANARI backend
- VTKm & VTKm-graph
- ANARI Volume Viewer
- Barney mutli-threaded renderer
- hayStack (Barney viewer)
- Barney's "BANARI" ANARI backend
- hayStack's "HANARI" ANARI backend
- PBRT code from Physically Based Rendering book
ParaView usage
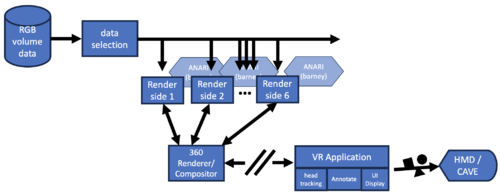
hayStack usage
The hayStack application uses the multi-GPU Barney rendering library
to display volumes with interactive controls of the opacity map. It is
intended to be a simple application that serves as a proof-of-concept
for the Barney renderer. There are a handful of command line options
and runtime inputs to know:
% ...
where
- 4@ — ??
- -ndg — ??
Runtime keyboard inputs:
- ! — dump a screenshot
- C — output the camera coordinates to the terminal shell
- E — (perhaps) jump camera to edge of data
- T — dump the current transfer function as "hayMaker.xf"
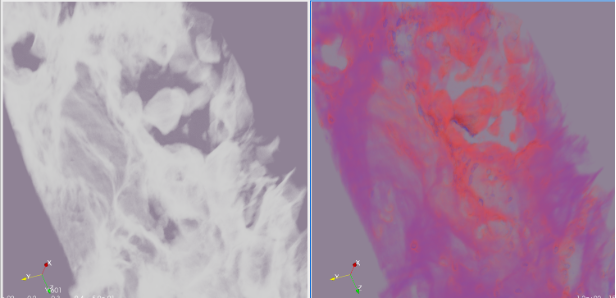
Example output (RGB tests):
(Original example -- single channel)
Process
Python Data Manipulation Scripts
Step 1: Tiff to Tiff converter & extractor
This program converts the data from the original Tiff compression scheme to an LZW compression scheme, and at the same time can extract a subvolume and/or reduce the data samples of the volume.
Presently all parameters are hard-coded in the script:
- input_path
- reduce — sub-sample amount of the selected sub-volume
- startAt — beginning of the range to extract (along the X? axis)
- extracTo — end of the range to extract (along the X? axis)
Step 2: process_skin-<val>.py
This program reads the LZW-compressed Tiff file from step 1, and first extracts the R, G & B channels from the data. Using the RGB values, additional color attributes are calculated that can be used as scalar values that represent particular components of the full RGB color. Finally, the data is written to a VTK ".vti" 3D-Image file.
There are (presently) two versions of this file, the first ("process_skin.py") was hard-coded to the specific parameters of the early conversion tests. The second ("process_skin-e300.py") is being transitioned into one that can handle more "generic" (to a degree) inputs.
In the future, I will also be outputting "raw" numeric data for use with tools that only deal in the bare-bones data.
The current (hard-coded) parameters are:
- extract — the size of the original data (used to determine R,G,B spacing)
- gamma — the exponential curvature filter to apply to the data